Welcome to the SeqDA-HLA_Influ Tutorial !
SeqDA-HLA_Influ is a model designed for predicting the binding of influenza-derived peptides to HLA class I proteins, with an architecture identical to SeqDA-HLA but fine-tuned using Influenza virus data. This tutorial will guide you through the steps necessary to use the peptide-HLA binding affinity prediction tool.
Step 1: Inputting Your Data
Enter the peptide sequences in the designated input field. Make sure the sequences are formatted correctly, following the examples provided on the Binding Affinity Prediction page.
Please ensure each peptide sequence is on a new line.
Example:
MEYDAVATT
CTELKLSDY
SRYWAIRTR
Step 2: Selecting HLA Types
Select the HLA types that correspond to your study. You can select multiple HLA types to see how different types bind to the same peptide. You can select up to 20 HLA types.
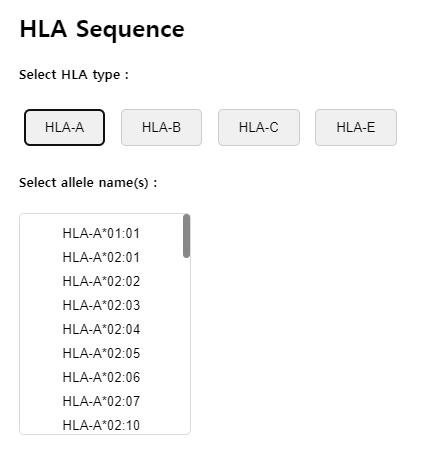
Step 3: Submitting and Interpreting Results
Click 'Submit' button to start the prediction. Once you submit your data, the results will be displayed on a new page. The results will include a list of binding affinities, helping you to understand the potential effectiveness of the peptide in triggering an immune response.
Go back to the Binding Affinity Prediction pages